Functions to do fast regression modelling. The functions return a tibble::tibble with statistics. Use plot() for an extensive model visualisation.
regression(x, ...)
# Default S3 method
regression(x, y = NULL, type = "lm", family = stats::gaussian, ...)
# S3 method for class 'data.frame'
regression(x, var1, var2 = NULL, type = "lm", family = stats::gaussian, ...)
# S3 method for class 'certestats_reg'
plot(x, ...)
# S3 method for class 'certestats_reg'
autoplot(object, ...)Arguments
Examples
runif(10) |> regression()
#> # A tibble: 2 × 5
#> term estimate std.error statistic p.value
#> * <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 7.81 1.98 3.93 0.00434
#> 2 x -5.39 4.11 -1.31 0.226
data.frame(x = 1:50, y = runif(50)) |>
regression(x, y)
#> # A tibble: 2 × 5
#> term estimate std.error statistic p.value
#> * <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 0.554 0.0853 6.50 0.0000000436
#> 2 x -0.00265 0.00291 -0.911 0.367
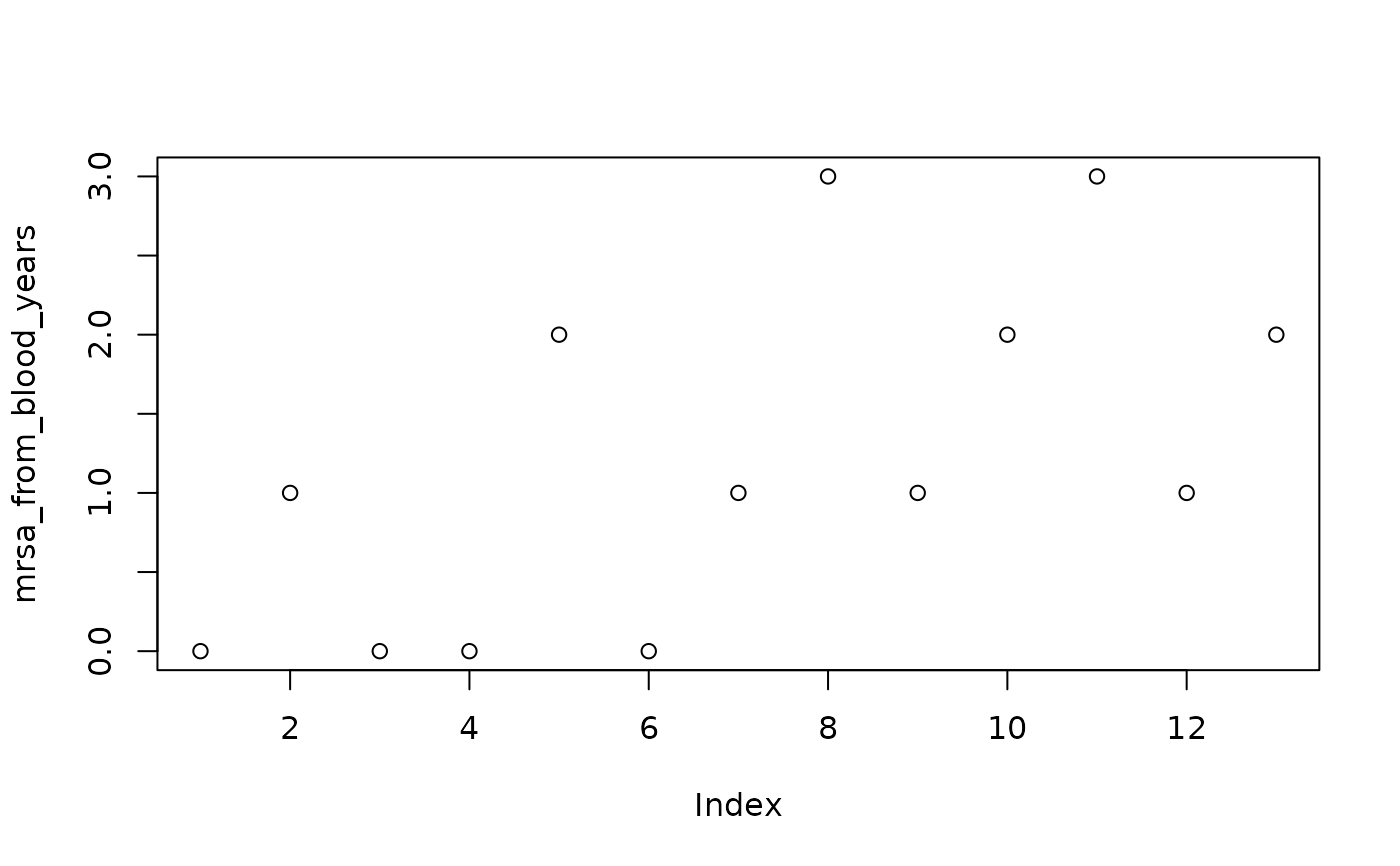
mrsa_from_blood_years <- c(0, 1, 0, 0, 2, 0, 1, 3, 1, 2, 3, 1, 2)
mrsa_from_blood_years |> plot()
 mrsa_from_blood_years |> regression()
#> # A tibble: 2 × 5
#> term estimate std.error statistic p.value
#> * <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 4.33 1.38 3.14 0.00946
#> 2 x 2.17 0.854 2.54 0.0277
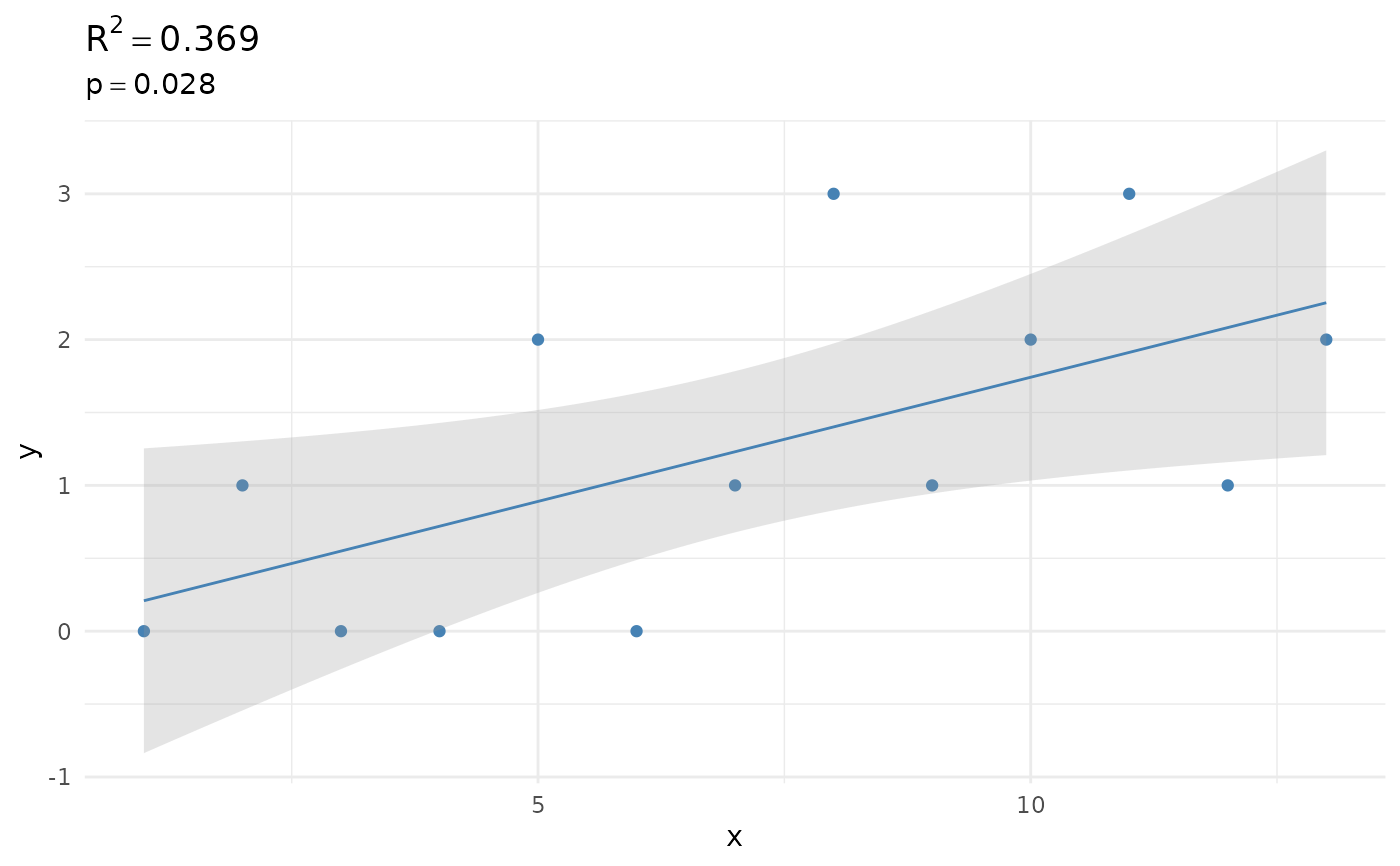
mrsa_from_blood_years |> regression() |> plot()
mrsa_from_blood_years |> regression()
#> # A tibble: 2 × 5
#> term estimate std.error statistic p.value
#> * <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 4.33 1.38 3.14 0.00946
#> 2 x 2.17 0.854 2.54 0.0277
mrsa_from_blood_years |> regression() |> plot()